Welcome to the webpage of the laboratory of Guillaume Lamoureux. Research in the lab is focused on applying theoretical and computational methods to fundamental problems in chemistry and biology.
We are interested in enzyme catalysis, membrane transport, and molecular recognition. Understanding these processes — and how they can be inhibited or enhanced — is at the core of many pressing issues in biology and medicine.
Our research is at the confluence of physical chemistry, computational biophysics, and structural biology. It relies on a solid understanding of molecular interactions and on a wide array of methods from statistical mechanics and machine learning. The laboratory is affiliated to the Center for Computational and Integrative Biology (CCIB).
Here are some active research projects in the laboratory:
Protein-protein interactions
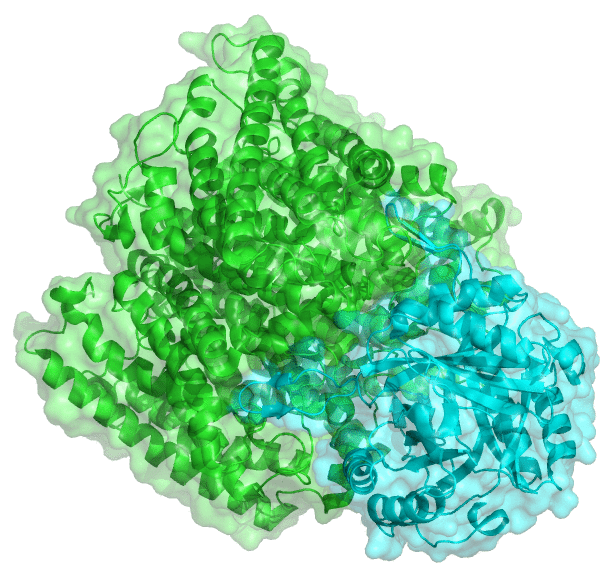
Proteins often acquire their biological activity by associating with one another and forming larger functional units. Finding out which two proteins are likely to interact is a difficult problem, since humans have around 20,000 protein-coding genes — creating a very large number of pairs of potential interaction partners. Moreover, this large network of interactions can be affected by mutations such as those occurring in cancer.
We are developing fast computer algorithms to predict protein-protein interactions and to infer which mutations are likely to perturb these interactions (and affect biological function). We are taking advantage of a number of recent developments in a field of machine learning called deep learning. Deep learning technologies are commonly used for automatic translation and for voice or image recognition. Treating the interaction of two proteins as a form of recognition — molecular recognition —, we are developing deep learning algorithms to predict whether any two proteins interact or not.
Computational enzymology and enzyme design
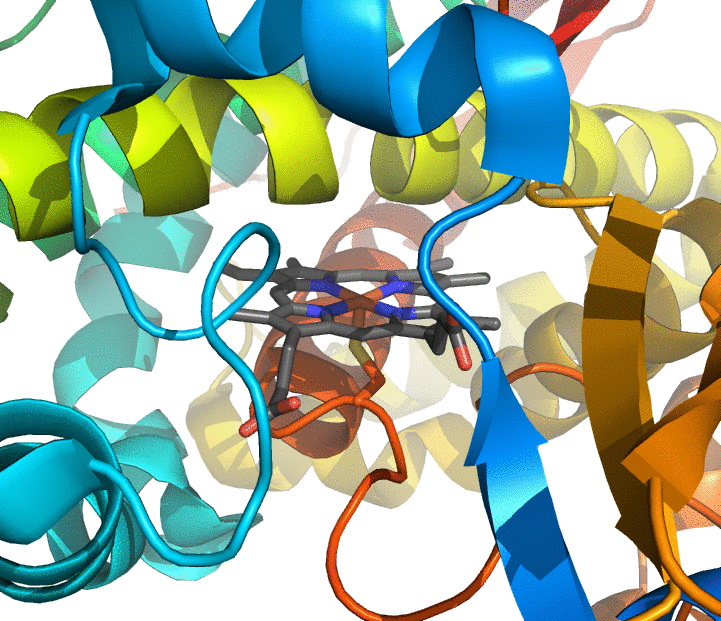
Computational enzymology is the study of enzymes and enzymatic reactions using molecular modeling and computer simulations. It aims primarily at describing the structure and relative energy of the short-lived states along an enzymatic reaction and at connecting them into an atomically-detailed kinetic picture. Because these short-lived states are usually not accessible through experimental techniques, it can provide new levels of insight to otherwise well-characterized reactions, and new interpretations to poorly-understood reactions.
We are developing methods for computational enzymology and applying them to enzymes containing metal atoms at their active sites (zinc, iron, etc.). Although about half of all known proteins contain at least one metal atom, no method is currently available to efficiently and reliably simulate chemical reactions in metalloenzymes. We are focusing on enzymes of medical and biotechnological interest, such as cytochrome P450s and zinc hydrolases.
Ion channels and transporters
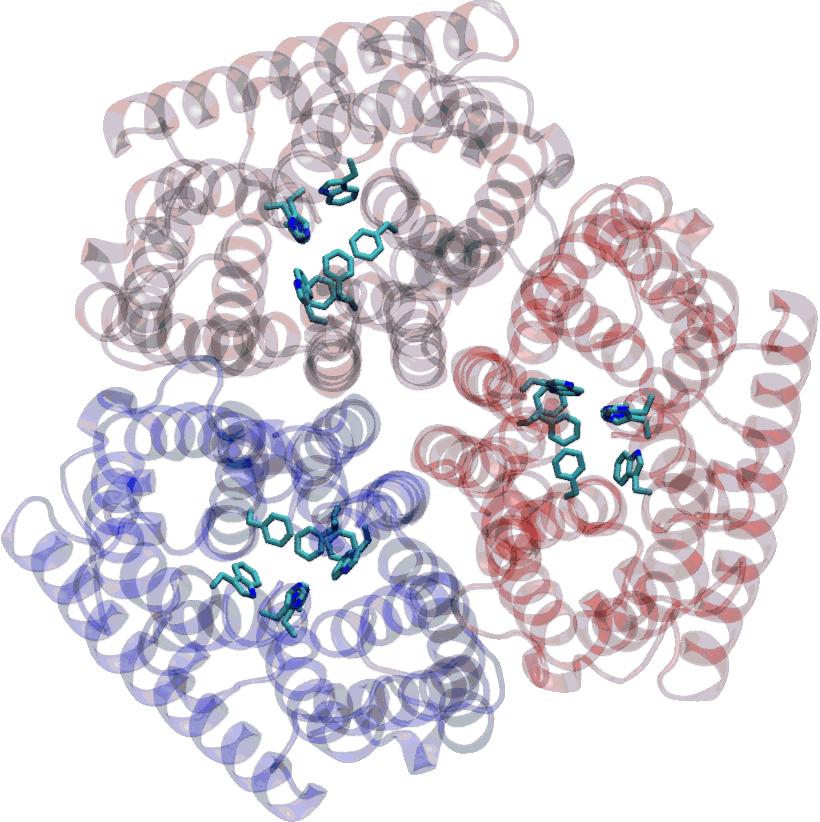
Living cells use a variety of channels and transporters to control molecular exchanges across their membranes. Although these transmembrane proteins are key to the understanding of cell physiology — and constitute major therapeutic targets for a great number of diseases — they are very challenging to study from a mechanistic point of view. Relatively few structures of transmembrane proteins are available, and those available tend to be difficult to interpret.
We investigate transport and permeation mechanisms in proteins using molecular modeling and molecular dynamics simulations. These techniques are particularly useful in revealing the structure and stability of intermediate states along the permeation pathway, which is precious information for rational drug design. In recent years, the laboratory has contributed to the understanding of the mechanism of Amt ammonium transporters, and is currently working on a number of ion channels of medical relevance.



